TurboPutative is a bioinformatic tool that streamlines the putative annotation process in Metabolomics, allowing data matrix handling and simplification, facilitating data visualization, useful information mining and, eventually, the prioritization of which candidate metabolites to investigate further. Click on this link to learn more about the usefulness and functioning of TurboPutative.
Using Web Server
It is possible to use TurboPutative from the browser in three simple steps. Click on this link to download an example input table.
1. Go to module selection
The webserver page must be accessed via the navigation bar or via the Try it now! button on the homepage.
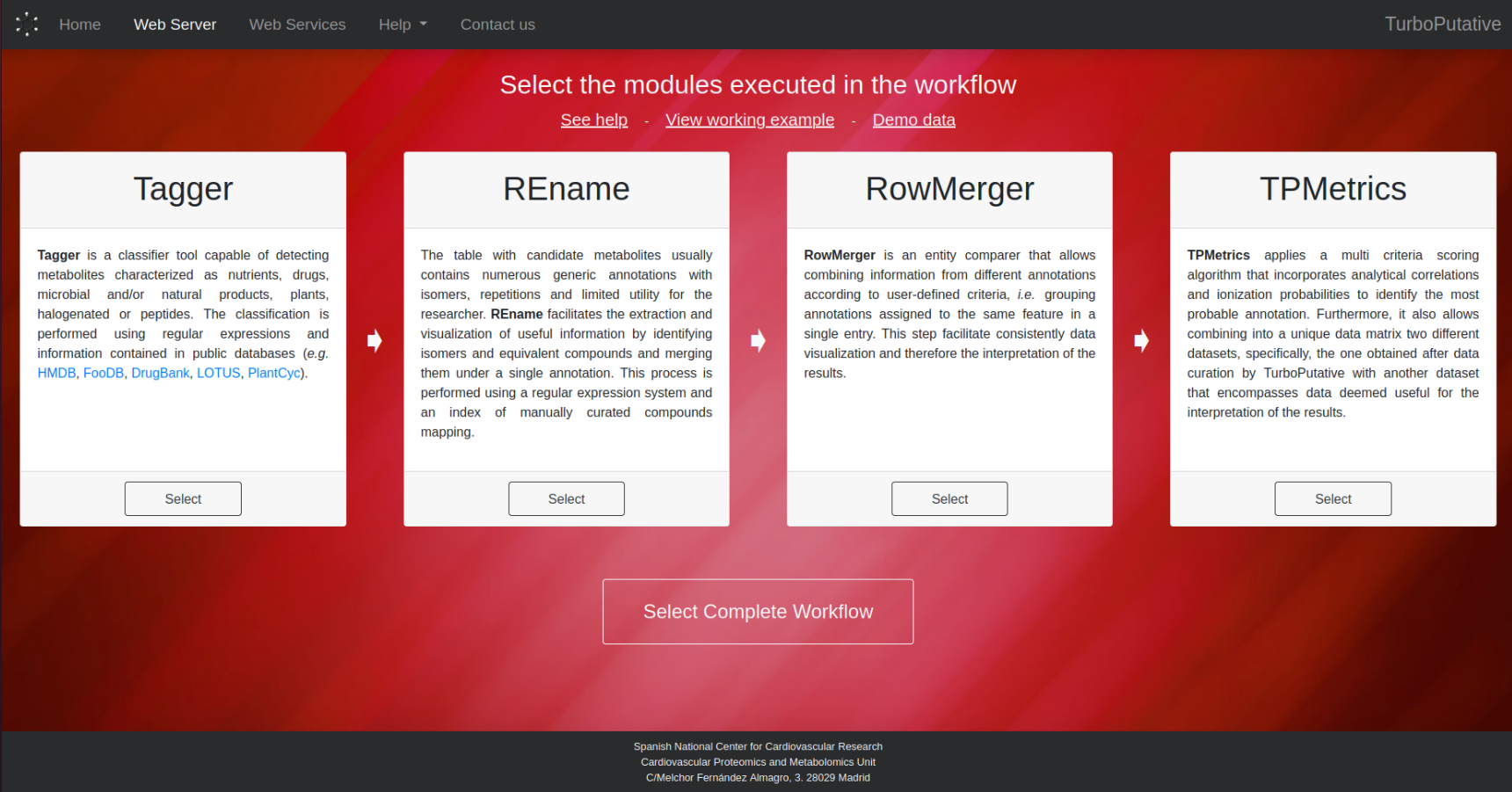
2. Select Modules
It must be selected the modules that are going to be run. The order in which the modules are run is Tagger > REname > RowMerger > TPMetrics, as indicated by the arrows. However, it is not necessary to select all the modules, as shown in the figure below.
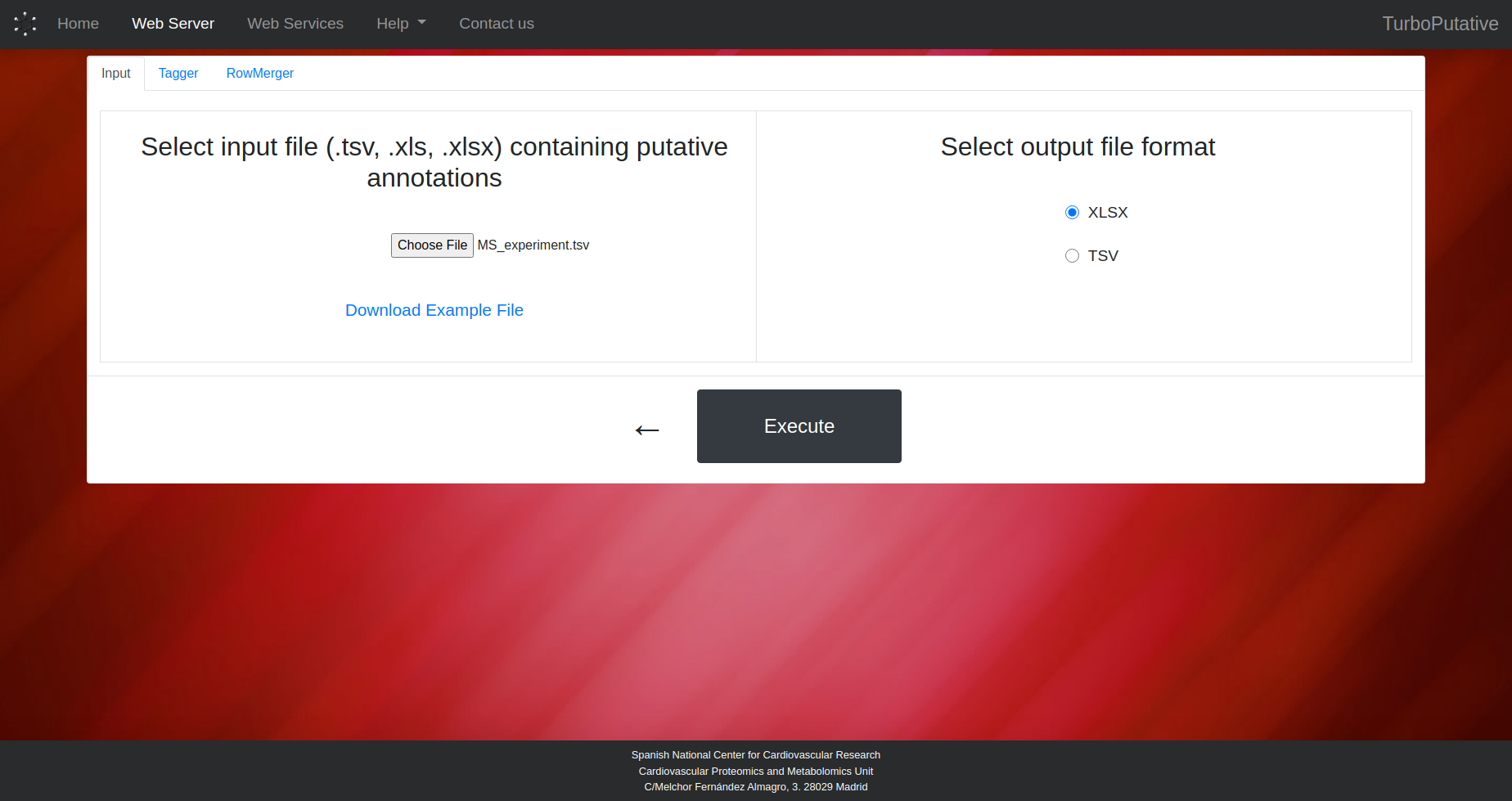
3. Customize Parameters
Next, upload the file with the putative annotations and configure the parameters of the selected modules. After finishing the configuration, press the Execute button to send the job to the server and access the upload page.
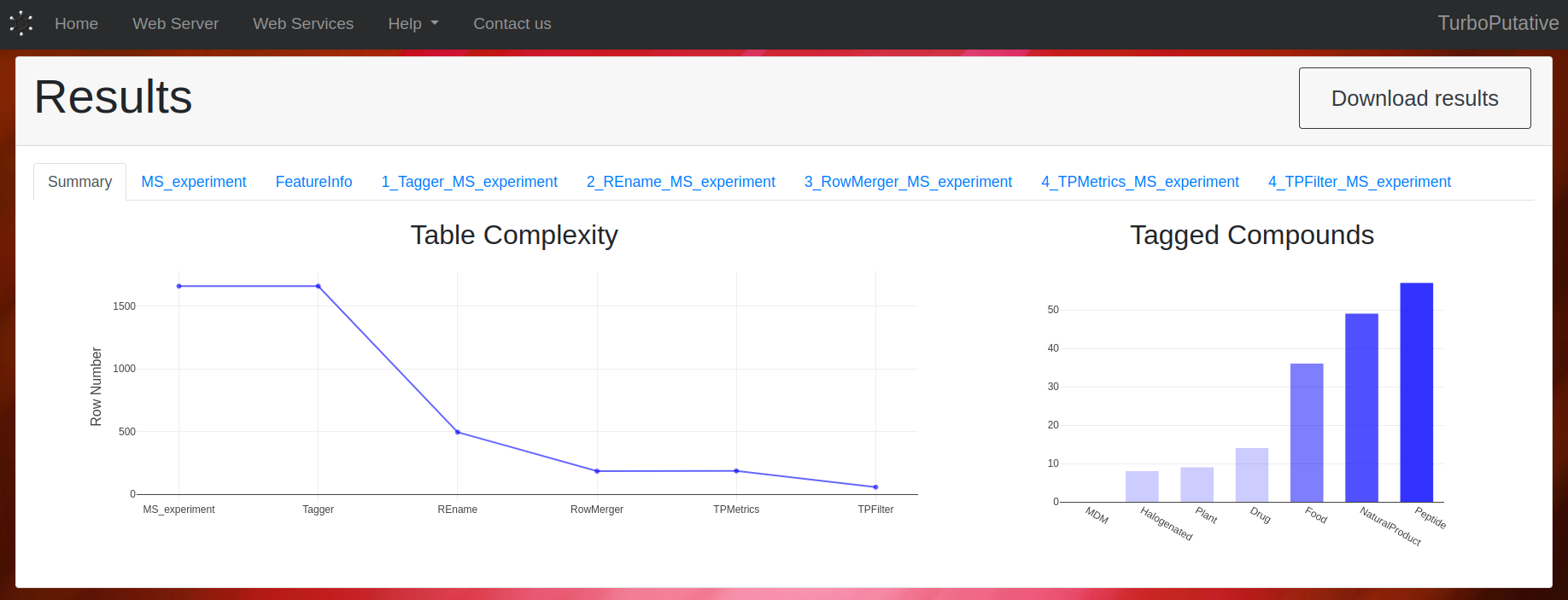
View Results
After the execution is finished, the main columns of the resulting tables will be displayed in the browser. From the browser it is possible to sort the rows and search for entries of interest. The complete tables can also be downloaded from the Download Results button.
Module Parameters
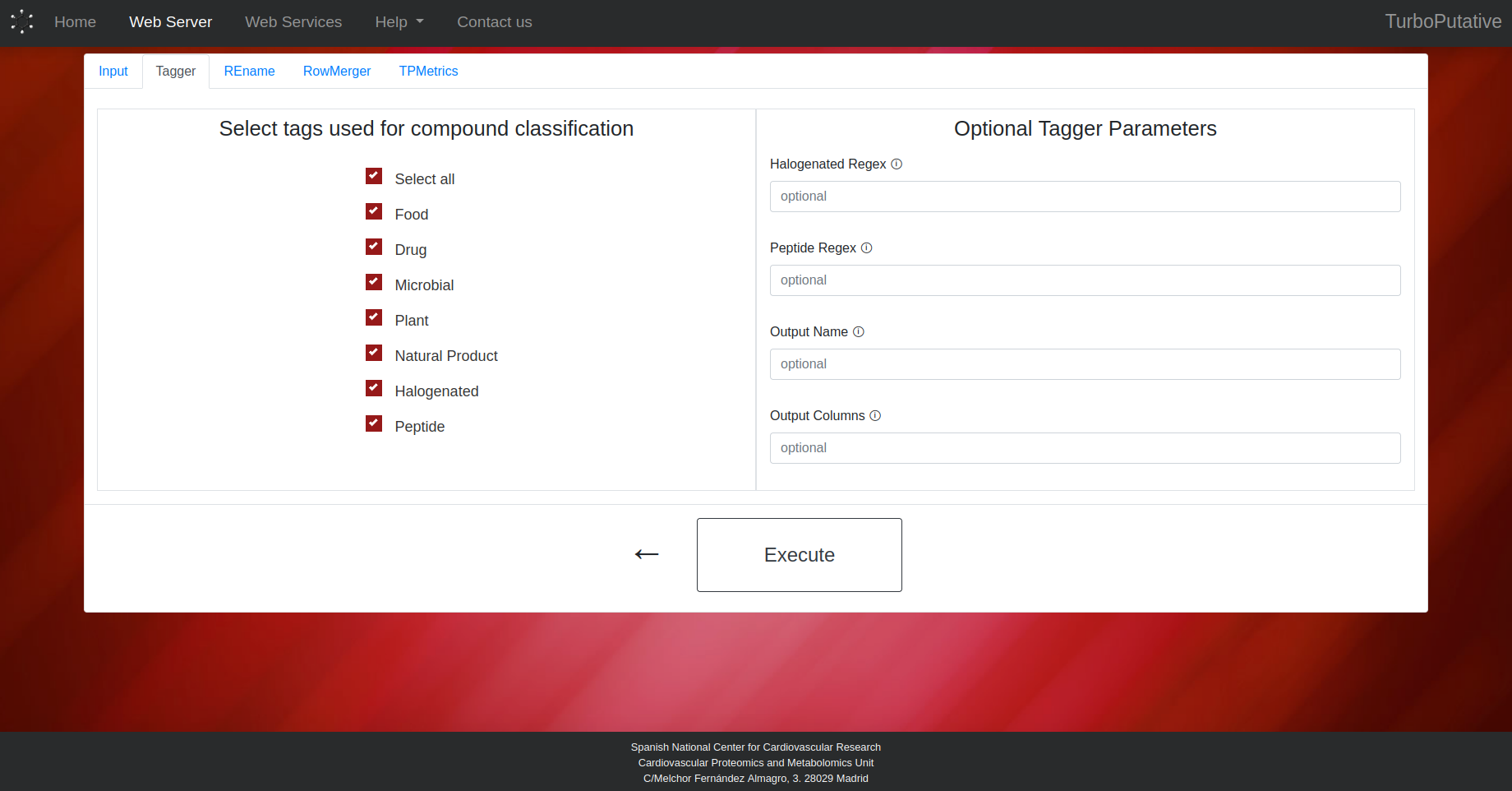
Tagger
The user can indicate the criteria (food, drug, microbial, plant, natural product, halogen and/or peptide) to be considered in the classification step. It is also possible to modify the following parameters:
- Halogenated Regex: Regular expresión applied on the name of the compounds to identify halogens. Default: "([Ff]luor(?!ene)|[Cc]hlor(?!ophyl)|[Bb]rom|[Ii]od)"
- Peptide Regex: Regular expresión applied on the name of the compounds to identify peptides.
Default:
"(?i)^(Ala|Arg|Asn|Asp|Cys|Gln|Glu|Gly|His|Ile|Leu|Lys|Met|Phe|Pro|Ser|Thr|Trp|Tyr|Val|[-\s,]){3,}$"
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “Tagger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
REname
- Remove Row: Regular expression that matches the name of the rows to be removed from the table. Default: “No compounds found for experimental mass”.
- Amino acid Separator: Regular expression that matches the characters that separate amino acids in a peptide. Thus, REname will be able to identify peptides with the same amino acid composition. Default: “\s”.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “REname_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
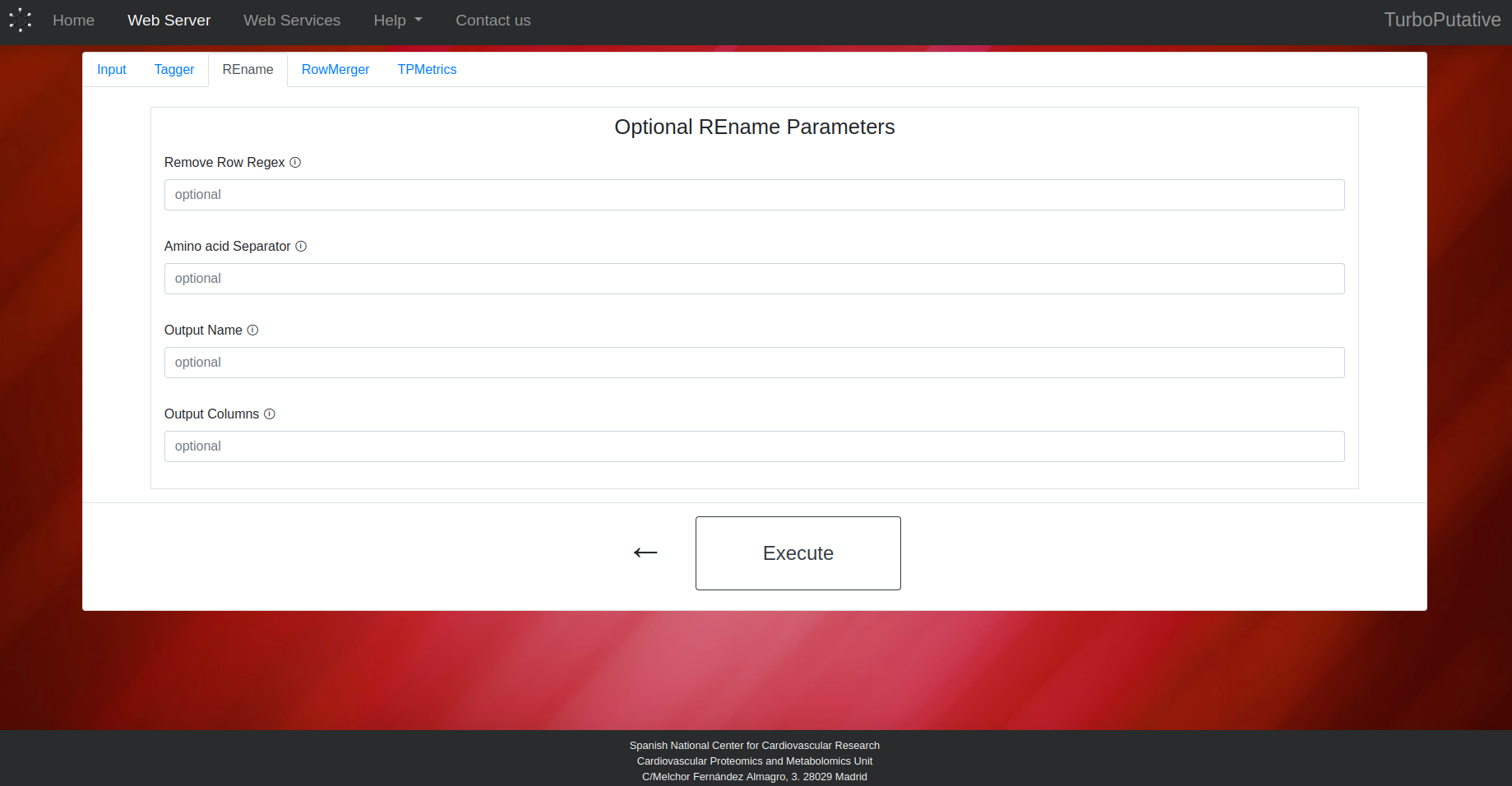
RowMerger
- Compared Columns: Name of the columns (separated by ‘,’) whose values will be compared when merging two rows. Two rows will be merged if they have the same values in these columns. Default: “Experimental mass, Adduct, mz Error (ppm)”.
- Conserved Columns: Name of the columns (separated by ‘,’) whose values will be preserved in the row resulting from the merge. The values of the merged rows will appear separated by " // ". By default, all columns will be conserved.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “RowMerger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
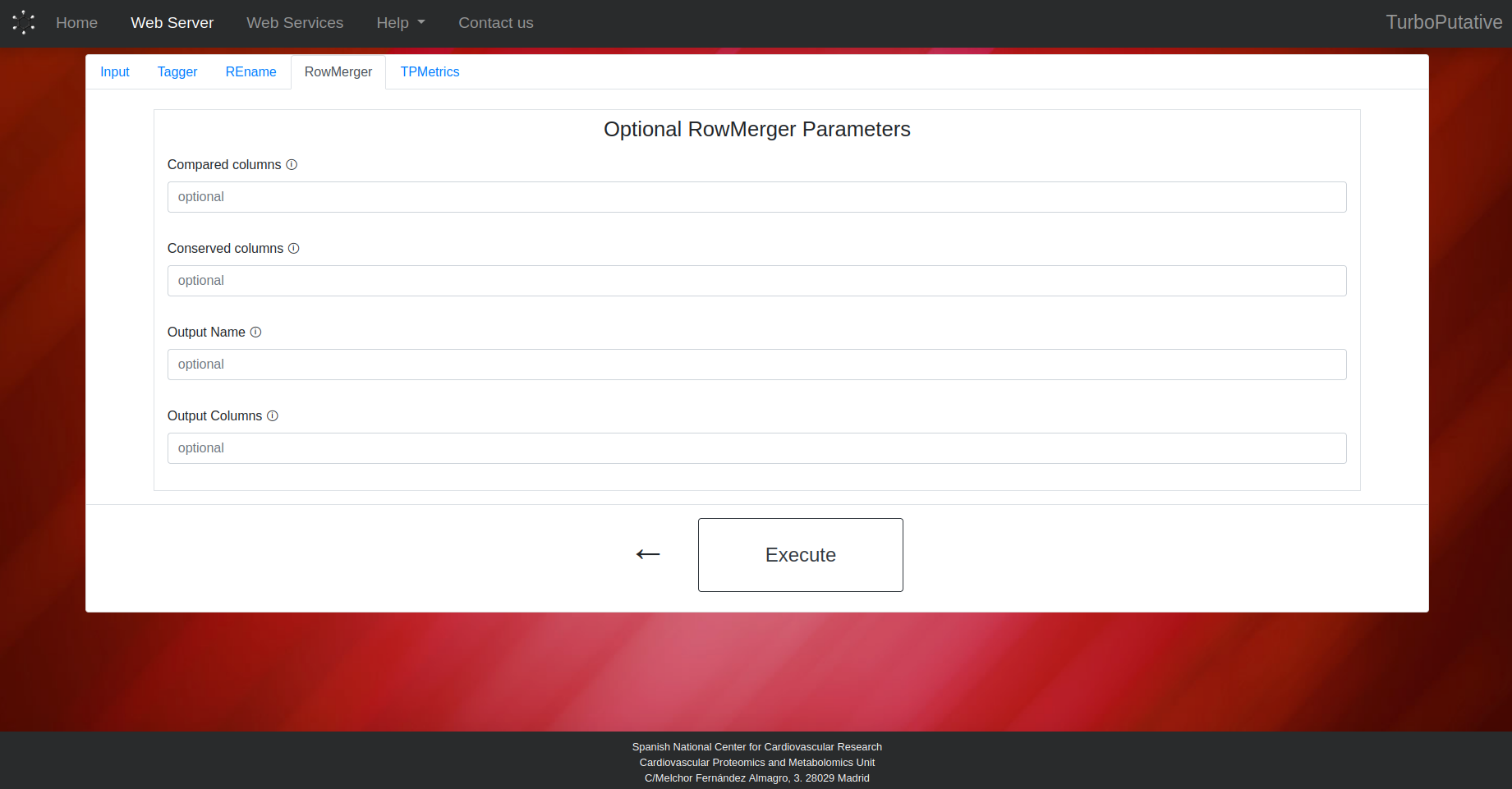
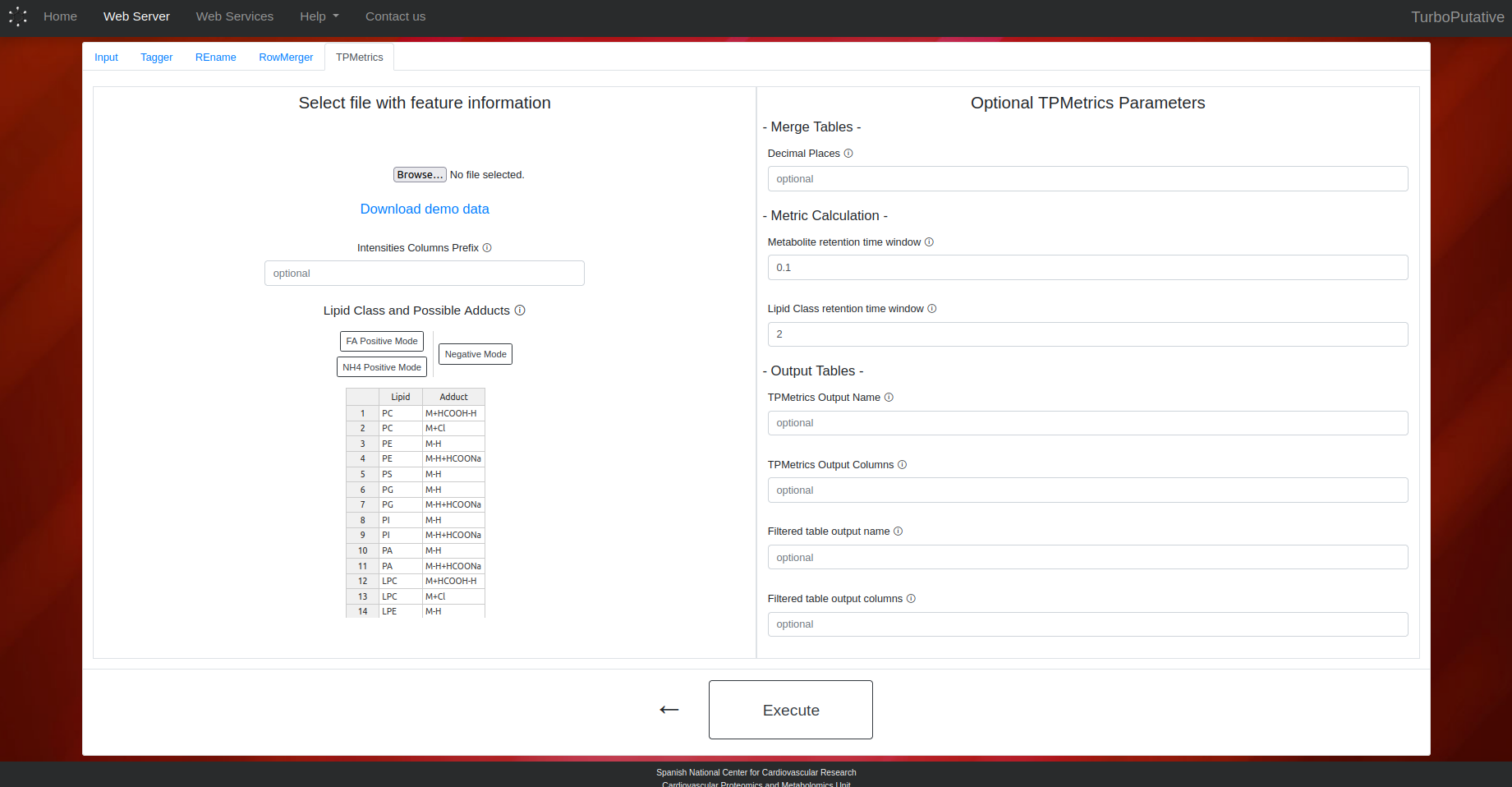
TPMetrics
Metric Calculation
- Intensities Columns Prefix: Pattern to find columns containing intensities. In demo data: C18_
- Lipid Class and Possible Adducts: Indicate the possible adducts associated with each lipid class for the score calculation. If a lipid class can have several adducts, the first ones will have a higher weight in the score calculation.
Merge Tables
- Decimal Places: Number of decimal places to which experimental mass of the features will be rounded. Default: 4.
Output Tables
- TPMetrics Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPMetrics_input-name.tsv”.
- TPMetrics Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
- Filtered Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPFilter_input-name.tsv”.
- Filtered Output Columns: Name of the columns present in the output filtered table. By default, all columns will appear in the output table.
Click on this link to download an example input table for TPMetrics.
It must be selected the modules that are going to be run. The order in which the modules are run is Tagger > REname > RowMerger > TPMetrics, as indicated by the arrows. However, it is not necessary to select all the modules, as shown in the figure below.

Next, upload the file with the putative annotations and configure the parameters of the selected modules. After finishing the configuration, press the Execute button to send the job to the server and access the upload page.

View Results
After the execution is finished, the main columns of the resulting tables will be displayed in the browser. From the browser it is possible to sort the rows and search for entries of interest. The complete tables can also be downloaded from the Download Results button.

Module Parameters
Tagger
The user can indicate the criteria (food, drug, microbial, plant, natural product, halogen and/or peptide) to be considered in the classification step. It is also possible to modify the following parameters:
- Halogenated Regex: Regular expresión applied on the name of the compounds to identify halogens. Default: "([Ff]luor(?!ene)|[Cc]hlor(?!ophyl)|[Bb]rom|[Ii]od)"
- Peptide Regex: Regular expresión applied on the name of the compounds to identify peptides.
Default:
"(?i)^(Ala|Arg|Asn|Asp|Cys|Gln|Glu|Gly|His|Ile|Leu|Lys|Met|Phe|Pro|Ser|Thr|Trp|Tyr|Val|[-\s,]){3,}$"
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “Tagger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

REname
- Remove Row: Regular expression that matches the name of the rows to be removed from the table. Default: “No compounds found for experimental mass”.
- Amino acid Separator: Regular expression that matches the characters that separate amino acids in a peptide. Thus, REname will be able to identify peptides with the same amino acid composition. Default: “\s”.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “REname_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

RowMerger
- Compared Columns: Name of the columns (separated by ‘,’) whose values will be compared when merging two rows. Two rows will be merged if they have the same values in these columns. Default: “Experimental mass, Adduct, mz Error (ppm)”.
- Conserved Columns: Name of the columns (separated by ‘,’) whose values will be preserved in the row resulting from the merge. The values of the merged rows will appear separated by " // ". By default, all columns will be conserved.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “RowMerger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

TPMetrics
Metric Calculation
- Intensities Columns Prefix: Pattern to find columns containing intensities. In demo data: C18_
- Lipid Class and Possible Adducts: Indicate the possible adducts associated with each lipid class for the score calculation. If a lipid class can have several adducts, the first ones will have a higher weight in the score calculation.
Merge Tables
- Decimal Places: Number of decimal places to which experimental mass of the features will be rounded. Default: 4.
Output Tables
- TPMetrics Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPMetrics_input-name.tsv”.
- TPMetrics Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
- Filtered Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPFilter_input-name.tsv”.
- Filtered Output Columns: Name of the columns present in the output filtered table. By default, all columns will appear in the output table.
Click on this link to download an example input table for TPMetrics.

The user can indicate the criteria (food, drug, microbial, plant, natural product, halogen and/or peptide) to be considered in the classification step. It is also possible to modify the following parameters:
- Halogenated Regex: Regular expresión applied on the name of the compounds to identify halogens. Default: "([Ff]luor(?!ene)|[Cc]hlor(?!ophyl)|[Bb]rom|[Ii]od)"
- Peptide Regex: Regular expresión applied on the name of the compounds to identify peptides.
Default:
"(?i)^(Ala|Arg|Asn|Asp|Cys|Gln|Glu|Gly|His|Ile|Leu|Lys|Met|Phe|Pro|Ser|Thr|Trp|Tyr|Val|[-\s,]){3,}$" - Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “Tagger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

- Remove Row: Regular expression that matches the name of the rows to be removed from the table. Default: “No compounds found for experimental mass”.
- Amino acid Separator: Regular expression that matches the characters that separate amino acids in a peptide. Thus, REname will be able to identify peptides with the same amino acid composition. Default: “\s”.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “REname_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

RowMerger
- Compared Columns: Name of the columns (separated by ‘,’) whose values will be compared when merging two rows. Two rows will be merged if they have the same values in these columns. Default: “Experimental mass, Adduct, mz Error (ppm)”.
- Conserved Columns: Name of the columns (separated by ‘,’) whose values will be preserved in the row resulting from the merge. The values of the merged rows will appear separated by " // ". By default, all columns will be conserved.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “RowMerger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

TPMetrics
Metric Calculation
- Intensities Columns Prefix: Pattern to find columns containing intensities. In demo data: C18_
- Lipid Class and Possible Adducts: Indicate the possible adducts associated with each lipid class for the score calculation. If a lipid class can have several adducts, the first ones will have a higher weight in the score calculation.
Merge Tables
- Decimal Places: Number of decimal places to which experimental mass of the features will be rounded. Default: 4.
Output Tables
- TPMetrics Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPMetrics_input-name.tsv”.
- TPMetrics Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
- Filtered Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPFilter_input-name.tsv”.
- Filtered Output Columns: Name of the columns present in the output filtered table. By default, all columns will appear in the output table.
Click on this link to download an example input table for TPMetrics.

- Compared Columns: Name of the columns (separated by ‘,’) whose values will be compared when merging two rows. Two rows will be merged if they have the same values in these columns. Default: “Experimental mass, Adduct, mz Error (ppm)”.
- Conserved Columns: Name of the columns (separated by ‘,’) whose values will be preserved in the row resulting from the merge. The values of the merged rows will appear separated by " // ". By default, all columns will be conserved.
- Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “RowMerger_input-name.tsv”.
- Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.

- Intensities Columns Prefix: Pattern to find columns containing intensities. In demo data: C18_
- Lipid Class and Possible Adducts: Indicate the possible adducts associated with each lipid class for the score calculation. If a lipid class can have several adducts, the first ones will have a higher weight in the score calculation.
- Decimal Places: Number of decimal places to which experimental mass of the features will be rounded. Default: 4.
- TPMetrics Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPMetrics_input-name.tsv”.
- TPMetrics Output Columns: Name of the columns present in the output table. By default, all columns will appear in the output table.
- Filtered Output Name: Name of the output file. Depending on the extension the output file format can change (.tsv, .xls, .xlsx). By default, the name of the output file will be “TPFilter_input-name.tsv”.
- Filtered Output Columns: Name of the columns present in the output filtered table. By default, all columns will appear in the output table.
Metric Calculation
Merge Tables
Output Tables
Click on this link to download an example input table for TPMetrics.
