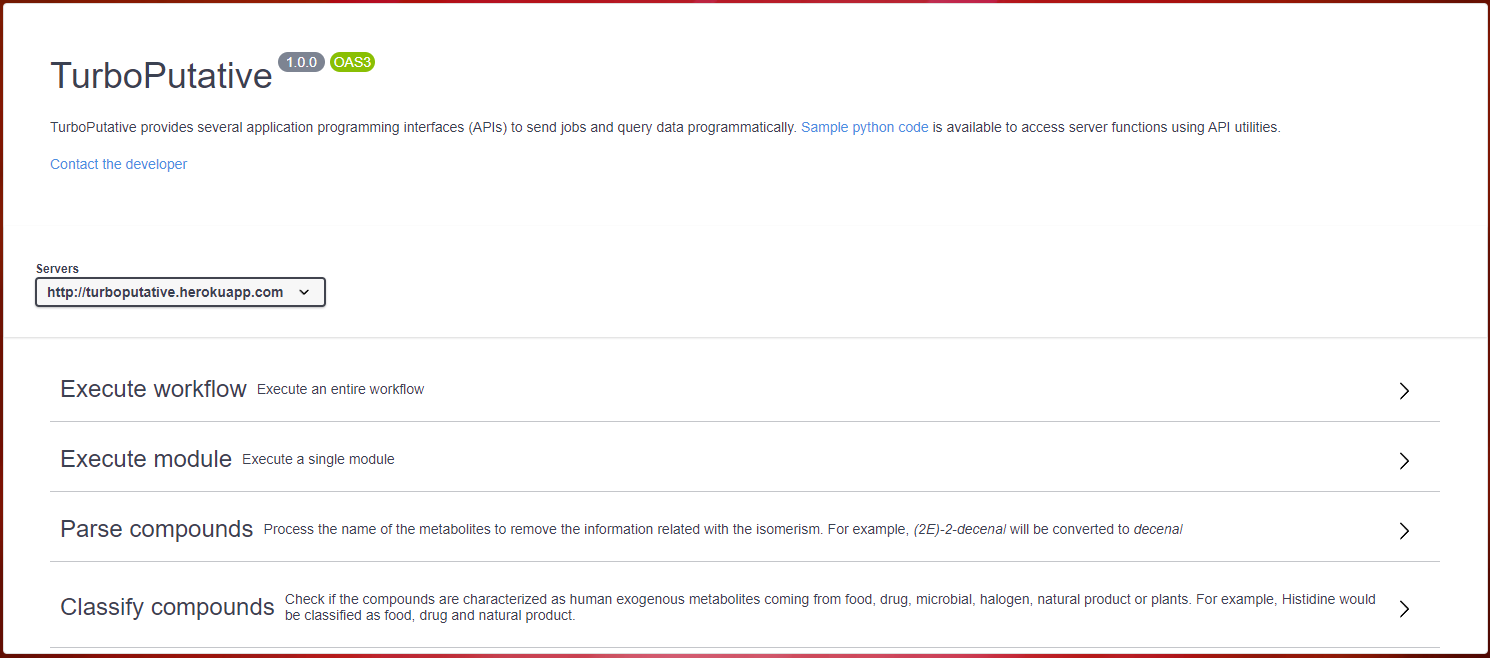
TurboPutative provides several application programming interfaces (APIs) to send jobs and query data programmatically. It is possible to make use of these utilities from the browser or by means of a Python script developed for this purpose. The four basic operations that can be carried out are the execution of a customized workflow, the execution of a specific module, the processing of the name of the compounds and their classification. Below, it is explained how to proceed in each case.
REST API from the Browser
To use the REST APIs from the browser, access the Web Services section from the top navigation bar. It will be found four sections that allow to carry out each of the operations mentioned above. In addition, each of the sections will show how to build a URL that allows the usage of the REST APIs from the terminal.
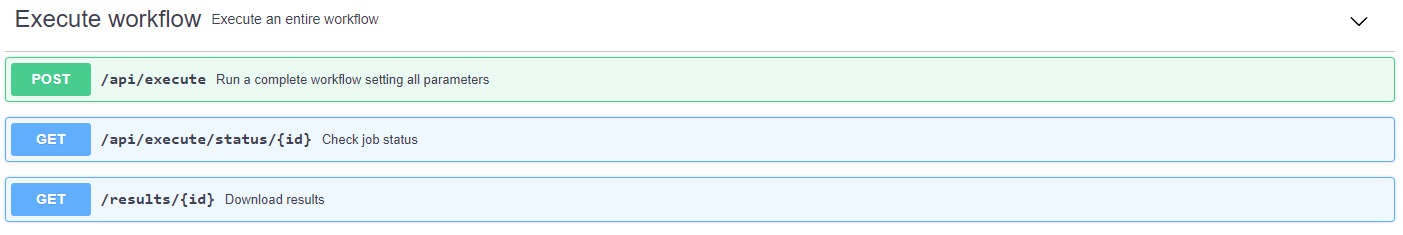
Execute workflow
The "Execute workflow" section allows to execute a complete workflow by customising each of the parameters. To do so, it is necessary to upload the table with the putative annotations (example file) and, optionally, a json file with the value of the parameters (example file). If the latter file is not uploaded, the default values will be used. Additionally, in case of using TPMetrics, it is necessary to indicate the table with the experimental mass of the features together with the information of interest (example file).
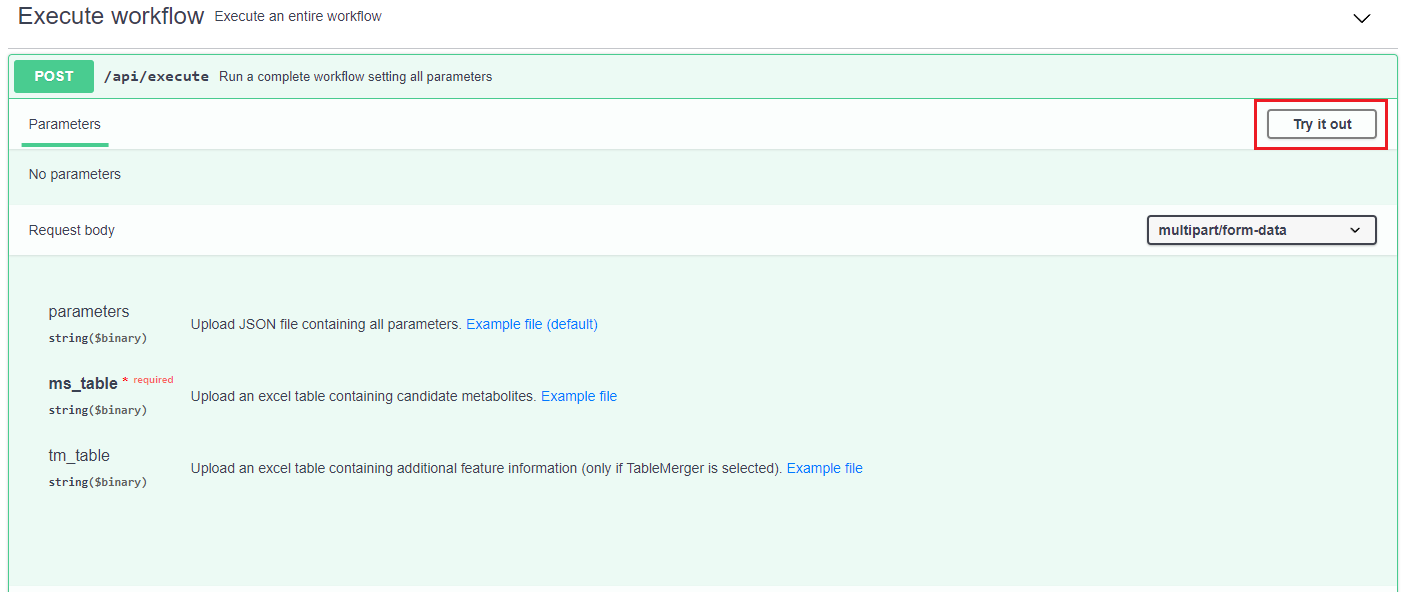
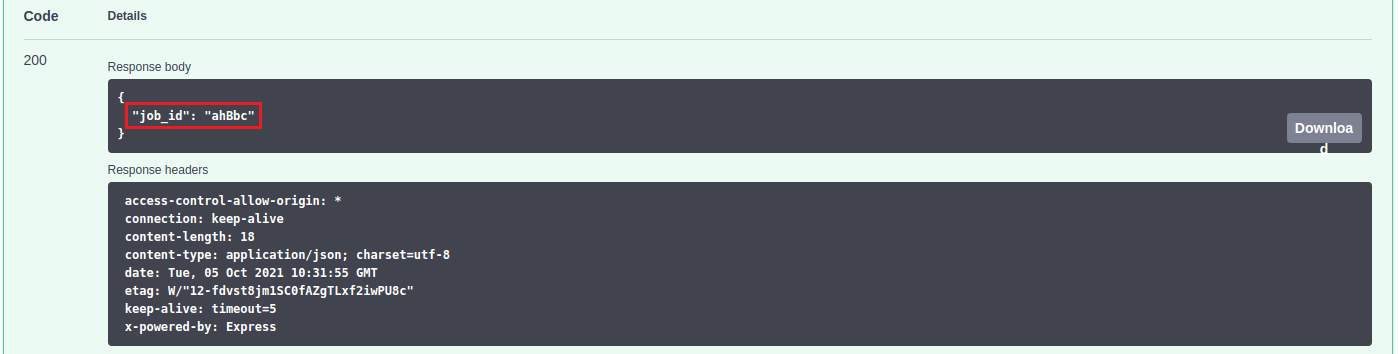
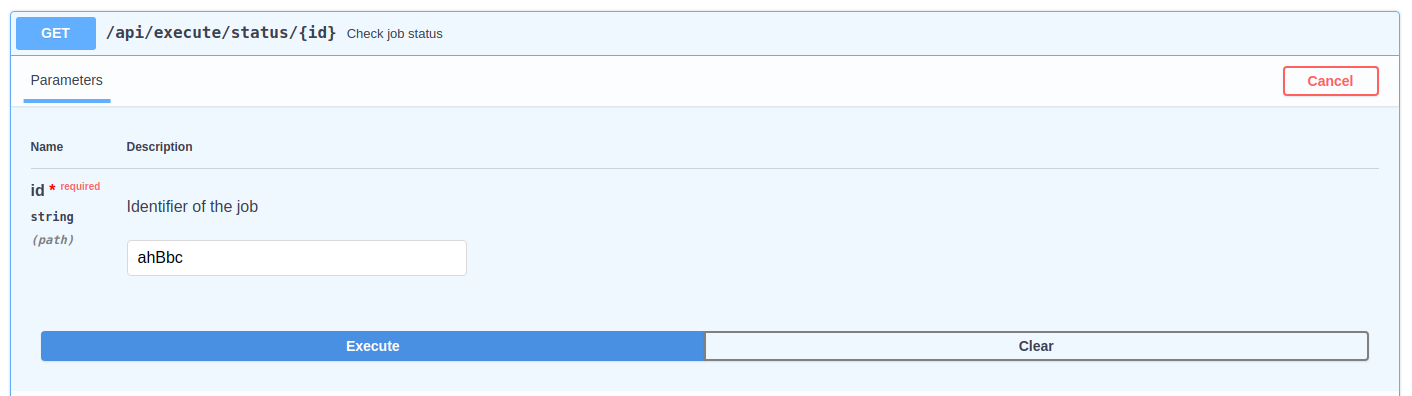
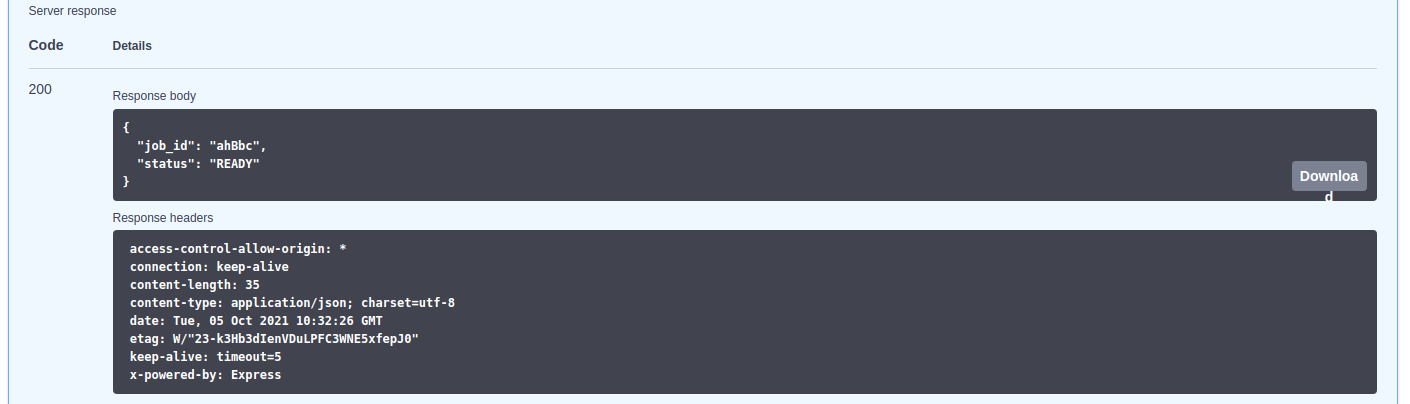
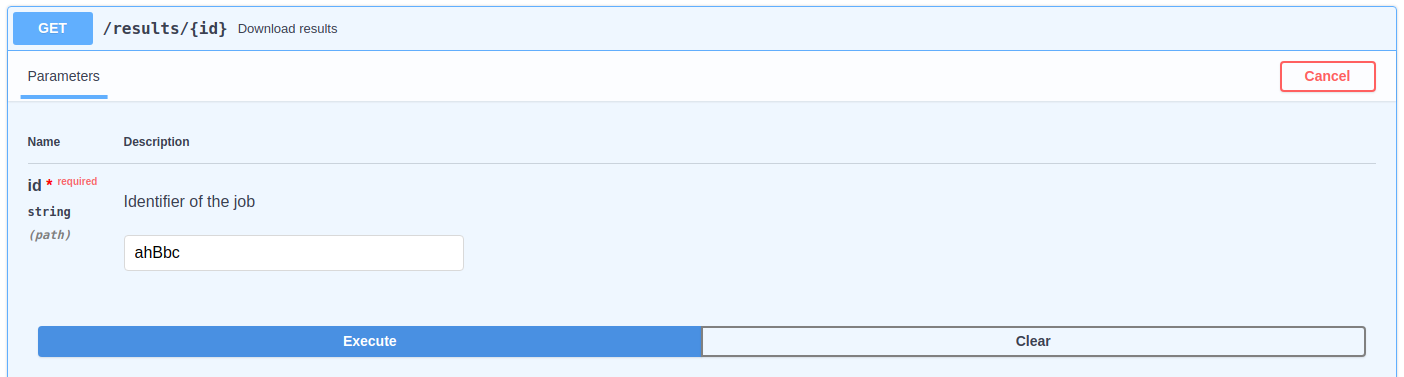
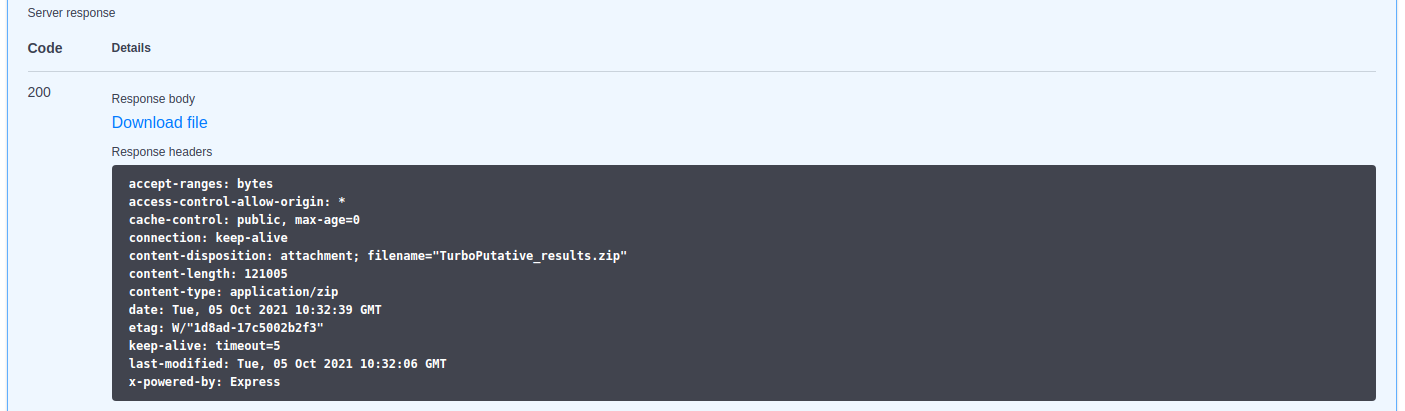
To check the status of the submitted job, it can be used the "status" paths shown in the above image. It must be indicated the job identifier and, when it is READY, it can be downloaded using the "result" path.
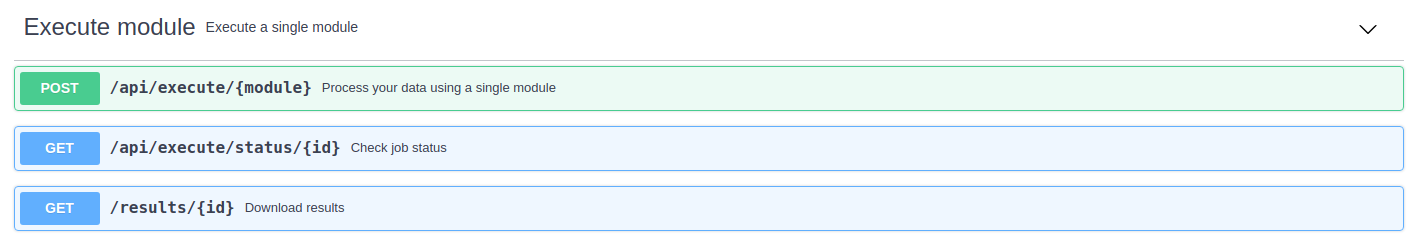
Execute module
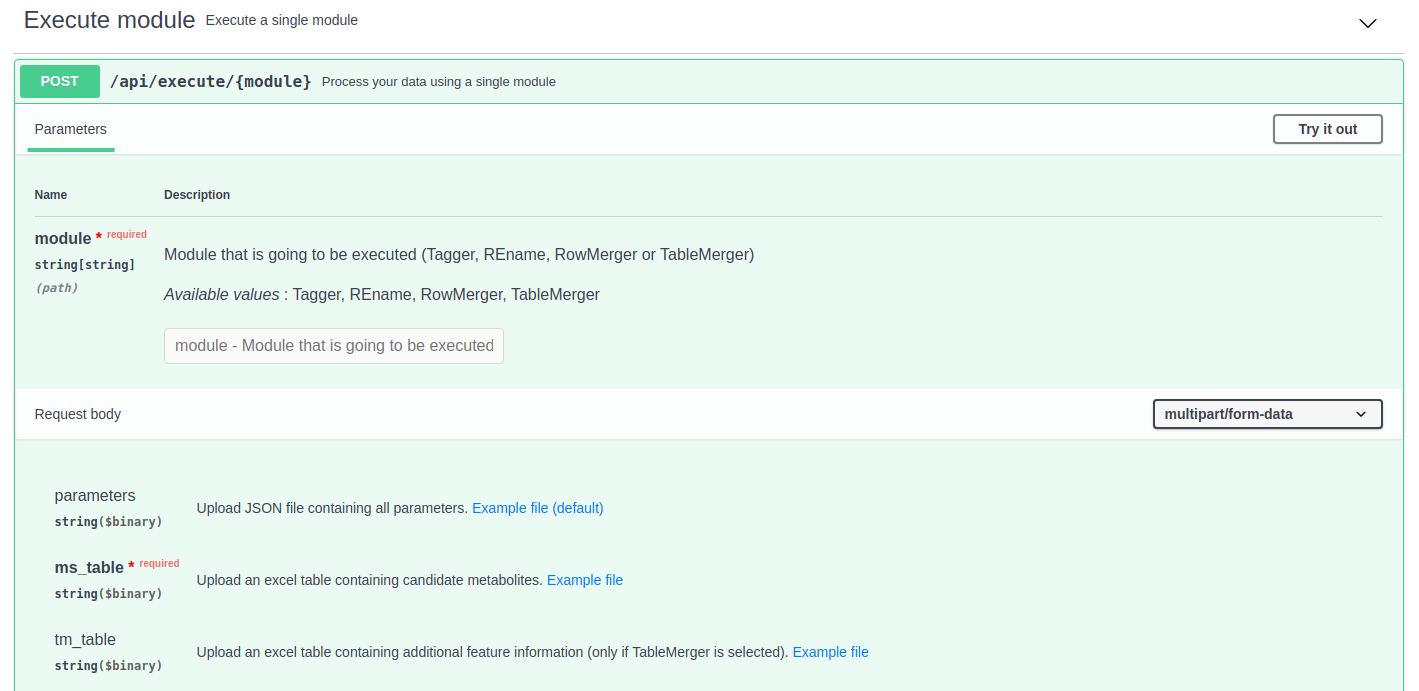
The "Execute module" section works in a similar way to the one described in the previous section. However, in this case, only a single module will be executed, which must be indicaded in one of the parameters.
Parse compounds
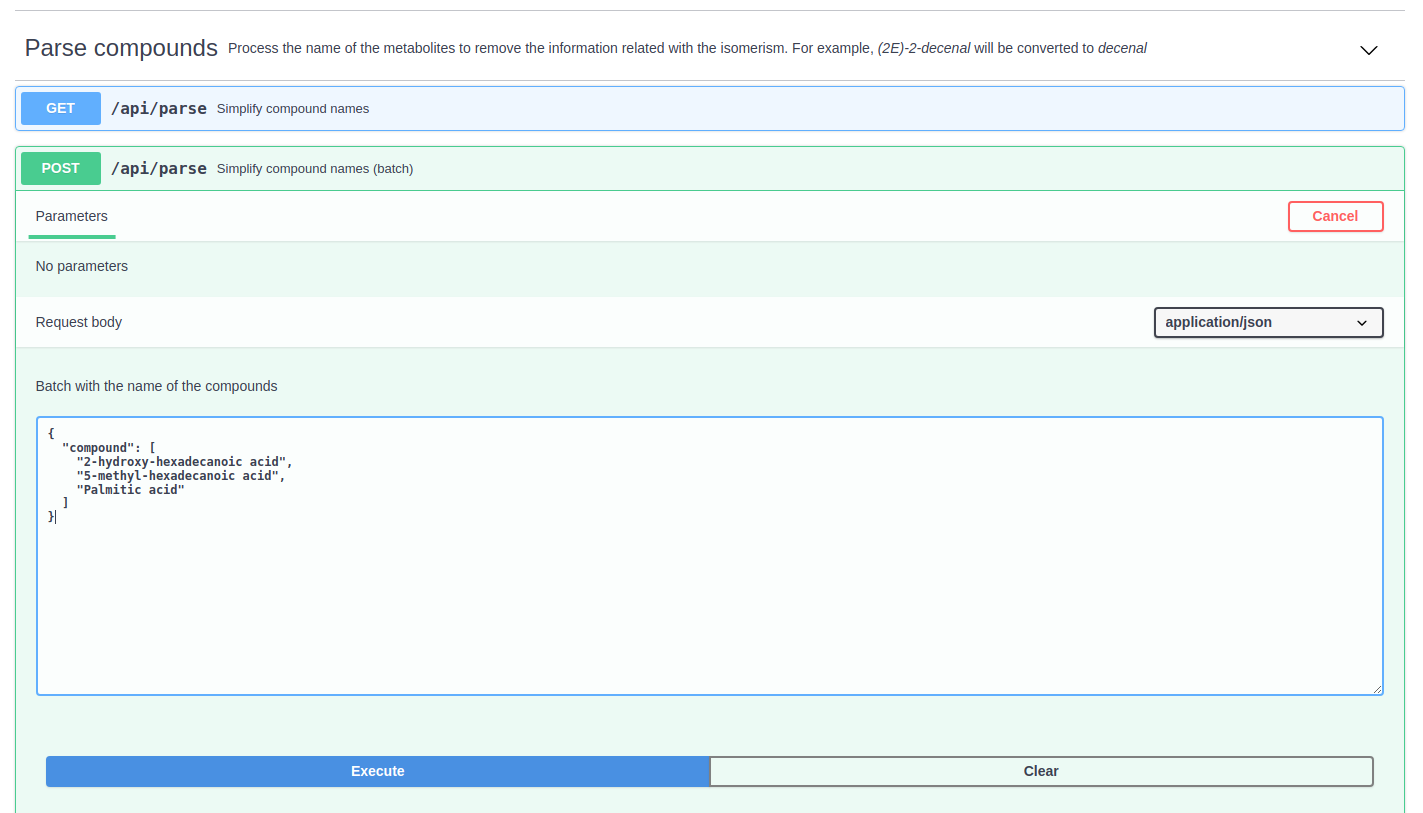
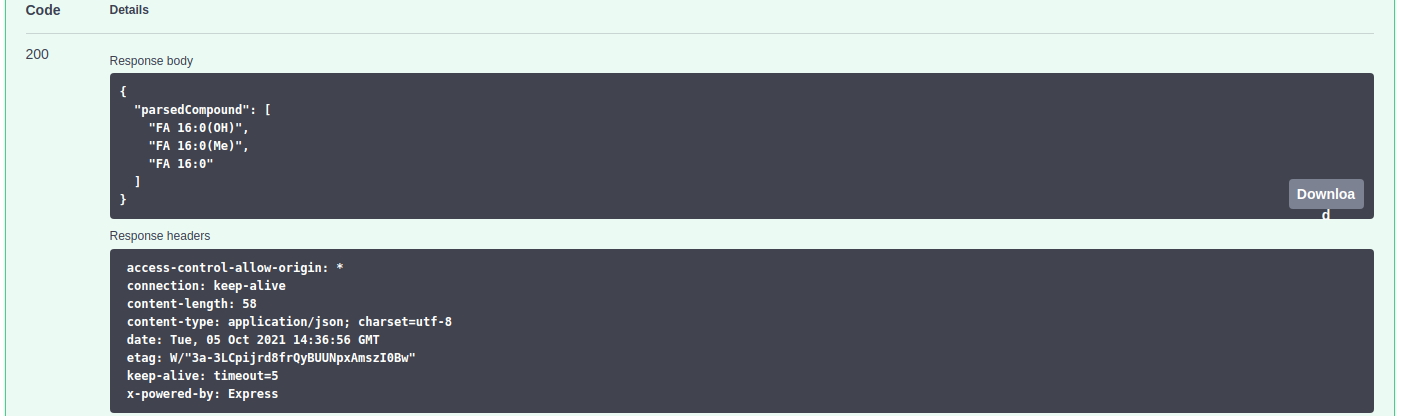
The "Parse compounds" option simplifies the name of the compounds to facilitate their classification and the identification of isomers. For example, the compound (2E)-2-decenal would be parsed to decenal.
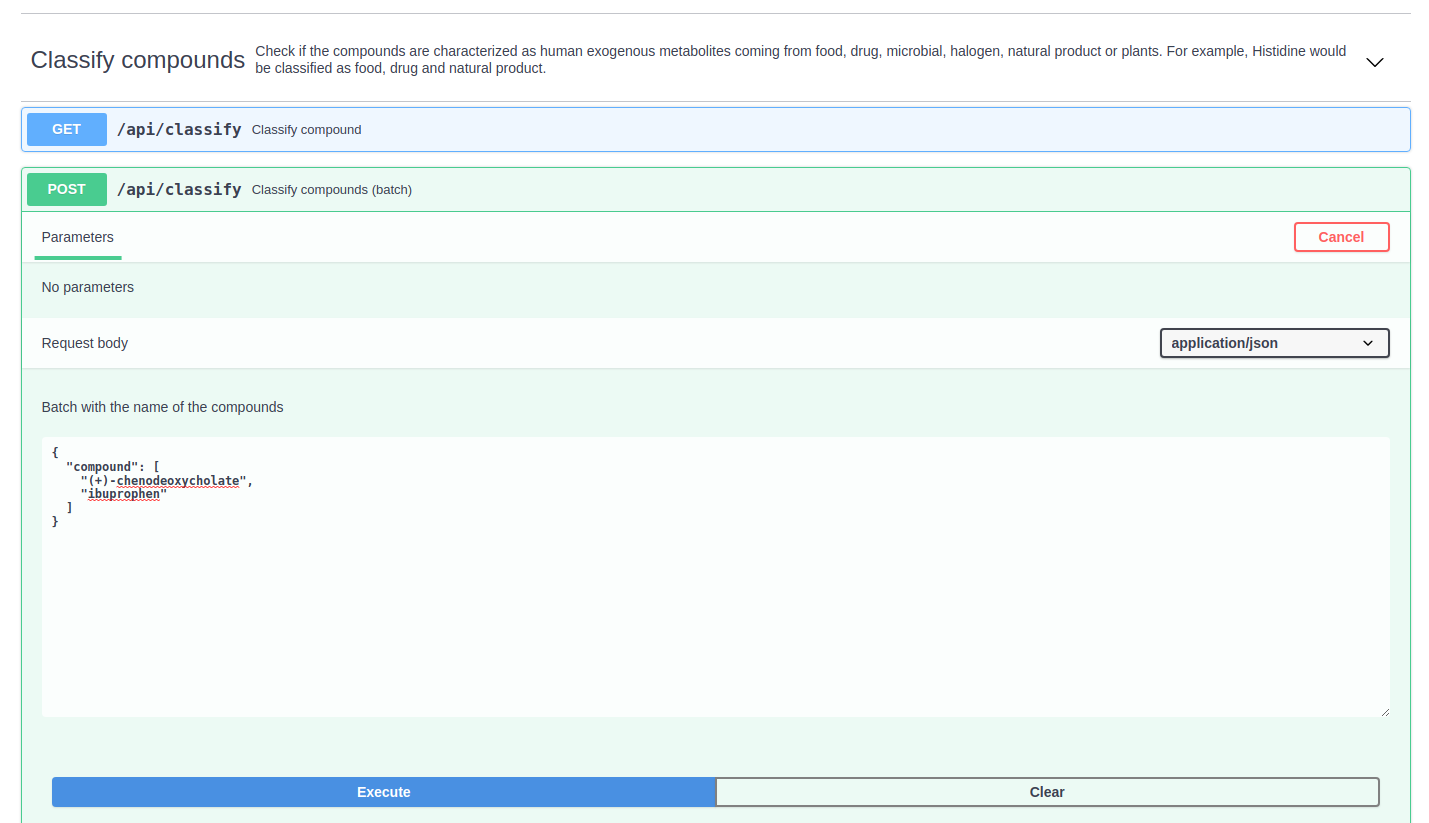
Classify compounds
The "Classify compounds" section is capable of detecting metabolites characterized as nutrients, drugs, microbiota-dependent metabolites, natural products, plants, halogens or peptides. The classification is performed using information contained in public databases (e.g. HMDB, DrugBank, PlantCyc, LOTUS, Metabolomics Data Explorer)
REST API from Python
It is possible to make use of the web services using the Python script that we make available to the user.
Execute workflow
It can be executed a complete workflow by customising each of the parameters. To do so, it is necessary to indicate the path to the table with the putative annotations (example file) and, optionally, a the path to a json file with the value of the parameters (example file). If the latter file is not uploaded, the default values will be used. Additionally, in case of using TPMetrics, it is necessary to indicate the path to the table with the experimental mass of the features together with the information of interest (example file).
$ python TurboPutative_API.py --workflow --msfile MS_experiment.tsv --tmfile FeatureInfo.tsv --param parameters.json
** Request workflow execution
** Job ID: O4iwC
$ python TurboPutative_API.py --status O4iwC
** Request job status
** Status (O4iwC):
{
'job_id': 'O4iwC',
'status': 'READY'
}
$ python TurboPutative_API.py --download O4iwC
** Request job results
** Results were downloaded and saved in /home/user/TurboPutative_results.zip
Execute module
The "Execute module" section works in a similar way to the one described in the previous section. A single module will be executed, which must be indicaded in one of the parameters.
$ python TurboPutative_API.py --workflow --module Tagger --msfile MS_experiment.tsv
** Request workflow execution
** Job ID: xQy1h
Parse compounds
The "Parse compounds" option simplifies the name of the compounds to facilitate their classification and the identification of isomers. For example, the compound (2E)-2-decenal would be parsed to decenal.
$ python TurboPutative_API.py --parse --compounds "(2E)-2-decenal" "hexadecanoic acid"
** Request compounds parsing
** Results:
{
'parsedCompounds': [
'decenal',
'FA 16:0'
]
}
** Results were saved in /home/user/parsedCompounds.json
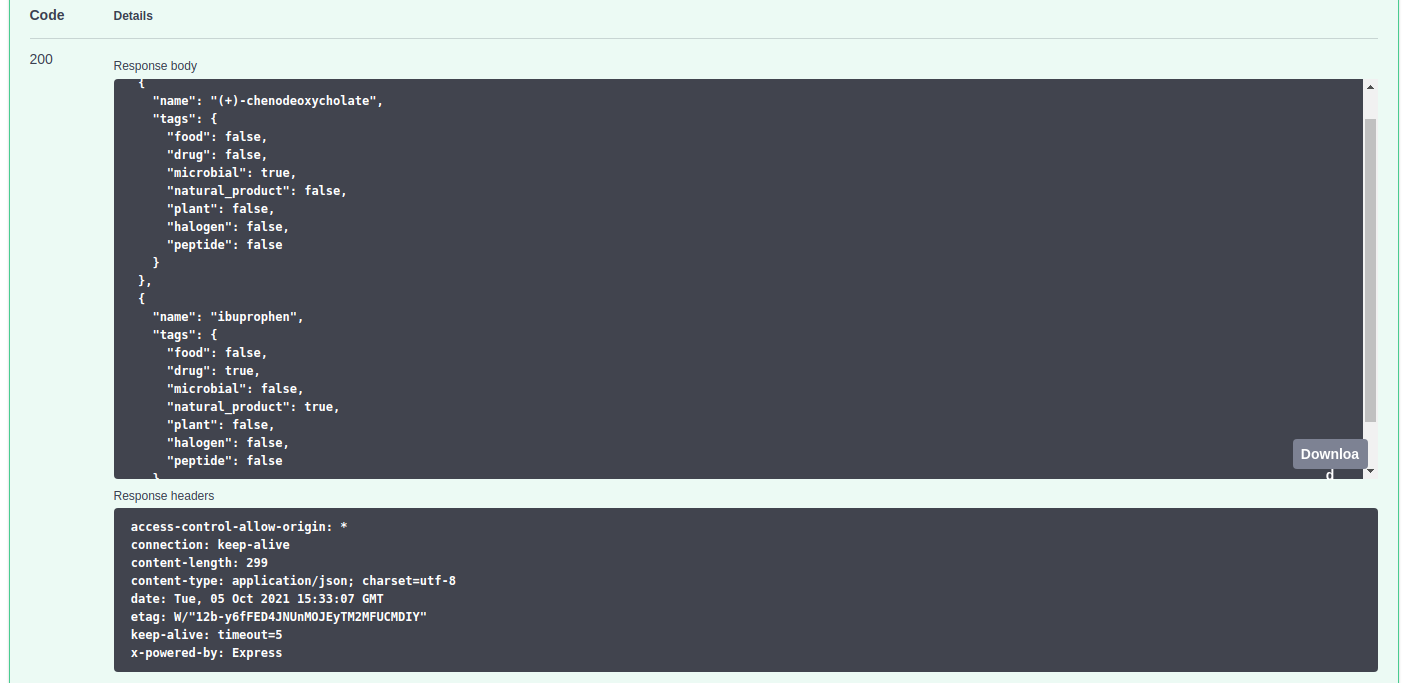
Classify compounds
API description
$ python TurboPutative_API.py --classify --compounds "chenodeoxycholate" "ibuprophen"
** Request compounds classification
** Results:
[
{
'name': 'chenodeoxycholate',
'tags': {
'food': False,
'drug': False,
'microbial': True,
'natural_product': False,
'plant': False,
'halogen': False,
'peptide': False
}
},
{
'name': 'ibuprophen',
'tags': {
'food': False,
'drug': True,
'microbial': False,
'natural_product': True,
'plant': False,
'halogen': False,
'peptide': False
}
}
]
** Results were saved in /home/user/classifiedCompounds.json